Tested with nebula sequencing data.
Credit: Most part of this article came from reddit author
Floedekartofler, link to original post. It's so helpful so I'm re-posting here.
This process is a bit involved since it's manual. I thought there would be open source programs available, but for some reason researchers in this field don't want to share. So instead I did it by hand. The advantage is that you don't need any command line tools. All you need is some high school biology knowledge.
Step 1: Look up the ABO gene in a genome browser
You can use the Nebula genome browser, but in case you like me didn't want to pay for the subscription you can also use the same genome browser at this address (https://igv.org/app/). Simply click tracks > local file and select the CRAM file as well as the CRAM index (CRAI). Next navigate to the ABO gene. The gene is located on chromosome 9 at position 133,250,401-133,275,201. Enter chr9:133,250,401-133,275,201 in the textbox next to the search bar. You should then see the ABO gene marked in blue on the refseq track and above it you'll see all your Nebula reads in the area and your genotype. If you click the cog next to the track, you can select the option to see all bases in your reads.
The ABO gene is reversed compared to the reference genome. Click the cog next to the top line of letters and select reverse, to see the reverse sequence instead. Make sure to also click the "highlight cursor" button at the top, so it will tell you which position you are hovering. Also turn on center line, so you can see which base you navigated to.
Recolic note: grey means your gene matched reference gene, colorful means yours doesn't match ref; You can choose reference data in
menu > Genome, most provider use GRCh38/hg38 as ref, but old data might use GRCh37/hg19.
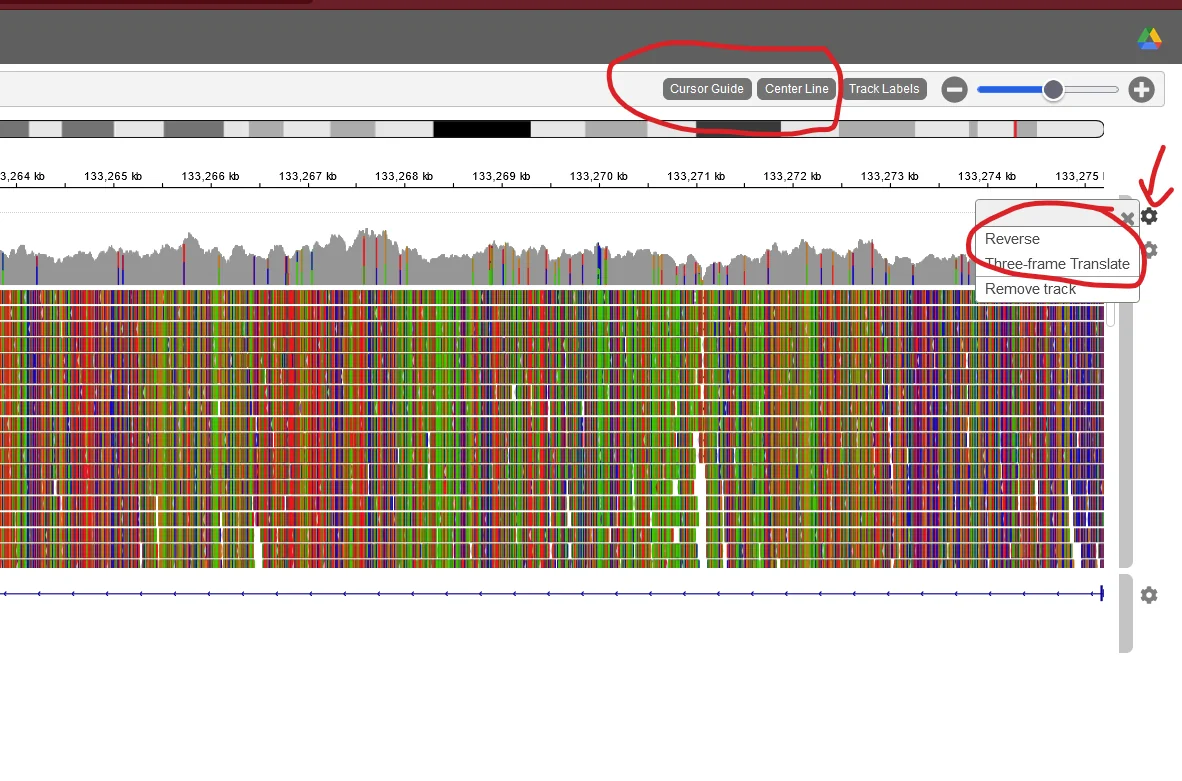
Turn on "cursor guide", "center line", and "three frame translate".
Type O or not type O
First navigate to position chr9:133,257,521-133,257,521. Most people with blood type O have a mutation here. The reference genome is blood type O, so if you have a G at position 133,257,523, a T at 133,257,522 and an A 133,257,521 you are type O.
If you on some of your reads see an I, it means that you have an extra base here compared to the reference genome. If you click the I it will tell you that this base is a C. This means that you are not type O.
If all of your reads have an I, you have two non-O alleles. If roughly half of them have an I you have one O and one non-O allele. If none of your reads have the I, you are homozygous for the deletion and thus blood type O.
There are other mutations that can give a type O blood type, but this one is the most common.
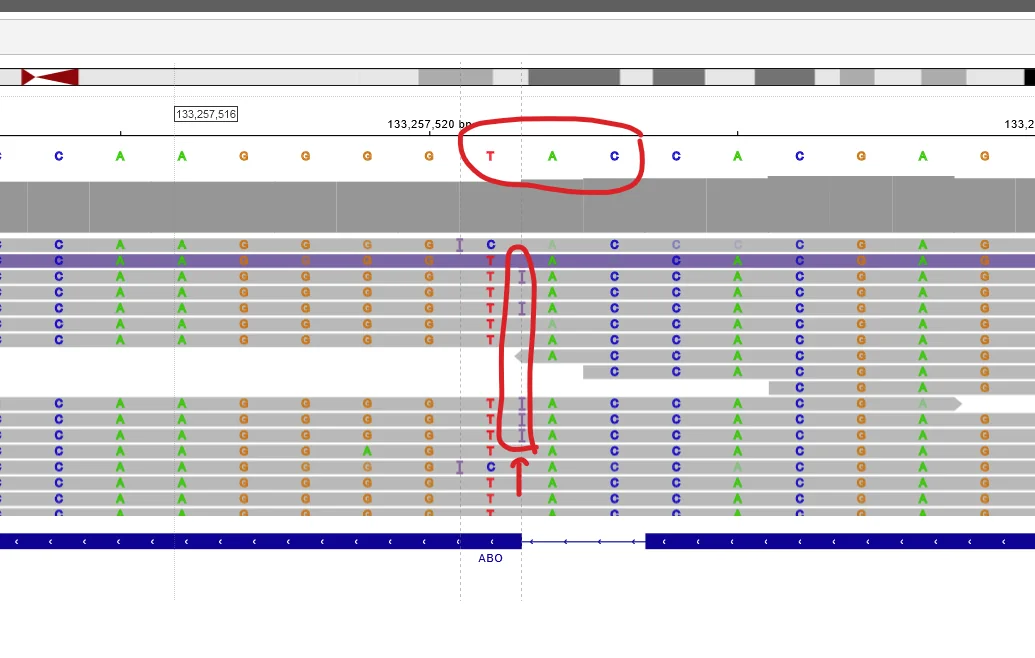
I am heterozygous for the deletion. This means I have one copy with the deletion (an O allele) and one copy without (a non-O allele). I can see I have a non O allele because I have some reads with an insertion. If I click the I it tells me the read was a C, meaning the gene had a G there. I also have some reads without the I with the sequence CAT. This is the deleted version.
Type A or B
If you were not type O you will need to figure out if you are type A or B. This is a bit more complex. Have a look at this paper https://pubmed.ncbi.nlm.nih.gov/12014997/. It has a nice figure that shows which variants correspond to which blood types. Here is an overview of which positions in the gene (text above letters on figure) correspond to which positions in the genome.
I recommend that you go through these positions and note your genotype. I just drew on top of the PDF. If I had a certain mutation I circled it and if I did not I crossed it. At the end my blood type was pretty easy to deduct. Remember that the gene is on the reverse strand. If you enabled reverse view (as mentioned earlier) the top line in the genome browser will have the correct base. You can verify the position by looking at the three bases in the figure. Remember to read from right to left in the genome.
However, the reads are not reversed, so when you look at the reads to determine genotype (especially important if you are heterozygous) remember to turn C into G, G into C, A into T and T into A. Also keep in mind that you have two alleles. So your genotype is the sum of two things from the table
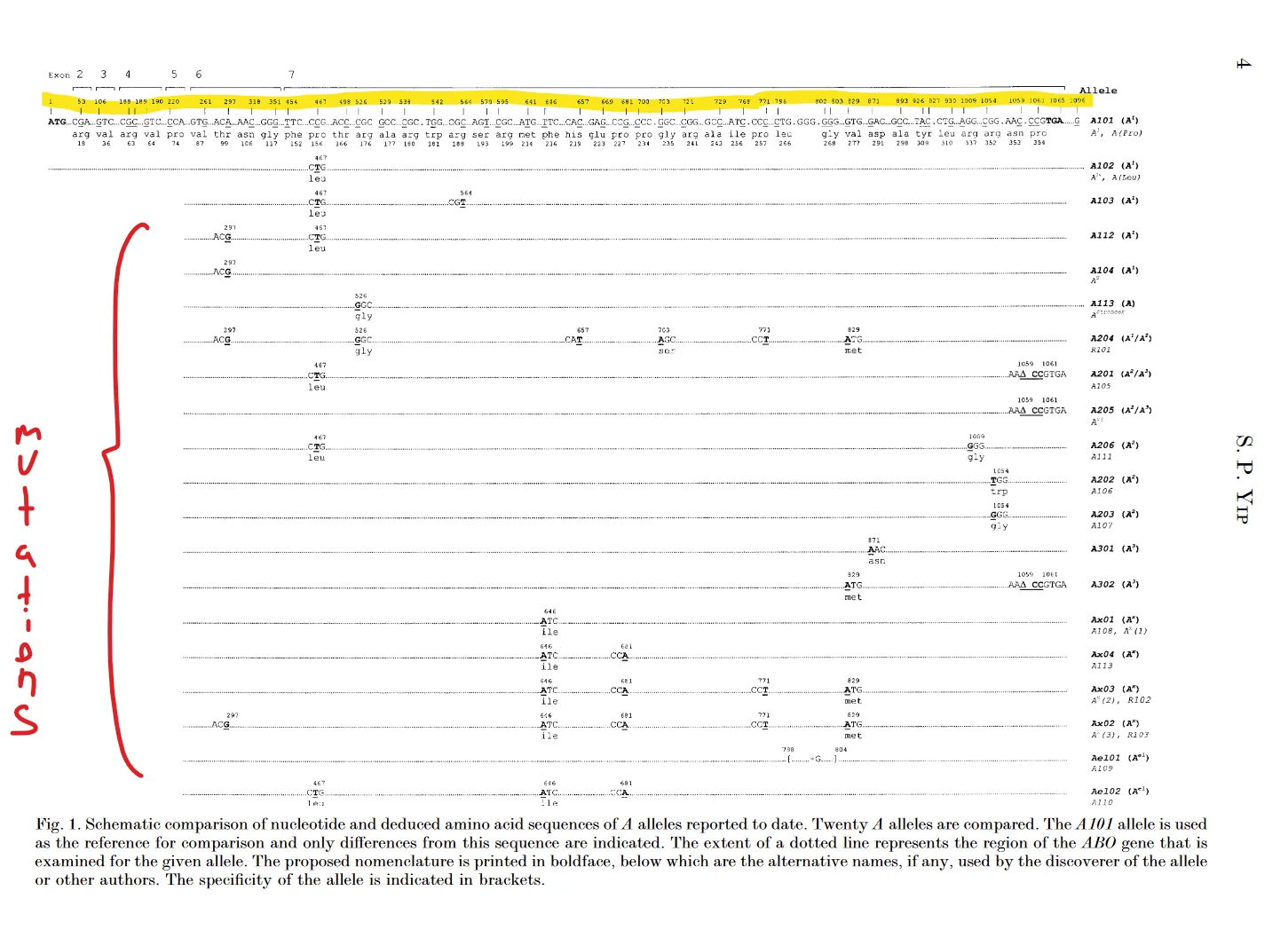
Look the yellow highlighted letters up in my table. Then enter chr9:pos-pos with pos being the position from the table. See if you have the mutations described.
1: 133,275,189
53: 133,262,144
106: 133,261,367
188: 133,259,834
189: 133,259,833
190: 133,259,832
220: 133,258,116
261: 133,257,521. This is the type O mutation discussed earlier.
297: 133,257,486
318: 133,257,465
351: 133,257,432
454: 133,256,277
467: 133,256,264
498: 133,256,233
526: 133,256,205
529: 133,256,202
538: 133,256,193
542: 133,256,189
564: 133,256,167
579: 133,256,152
595: 133,256,136
641: 133,256,090
646: 133,256,085
657: 133,256,074
669: 133,256,062
681: 133,256,050
700: 133,256,031
703: 133,256,028
721: 133,256,010
729: 133,256,002
768: 133,255,963
771: 133,255,960
796: 133,255,935
802: 133,255,929
803: 133,255,928
829: 133,255,902
871: 133,255,860
893: 133,255,838
926: 133,255,805
927: 133,255,804
930: 133,255,801
1009: 133,255,722
1054: 133,255,677
1059: 133,255,672
1061: 133,255,670
Recolic Appendix: RhD pos or neg
Additionally, I tried to learn my RhD pos/neg type but didn't found much useful info, so I asked GPT. It seems legit, works perfectly for me, so I'm sharing here for ref.
Disclaimer: This part is AI generated. It could be inaccurate!
| SNP | GRCh38 coordinate | REF base | Rh+ typical | Rh– typical |
|-------------|---------------------|----------|-------------|-------------|
| rs7853989 | chr1:25,694,681 | A | A/A | G/G |
| rs8176722 | chr1:25,667,747 | C | C/C | T/T |
| rs8176746 | chr1:25,681,029 | T | T/T | C/C |
| rs590787 | chr1:25,688,453 | G | G/G | A/A |
| RHD gene | chr1:25,570,000-25,690,000 | Exists | Exists | Deleted |
Leave a Reply